A Preliminary Phylogenetic Analysis of ITS nrDNA Sequences in Water-lilies
-
摘要: 睡莲类植物是目前植物分子系统学与进化研究的一个重要类群.核基因组的ITS区是核核糖体(nrDNA) 转录单位的一部分.测定和分析了5个属的7种睡莲类植物的核核糖体ITS序列, 并与GenBank中提取的相关序列进行了组合分析, 初步获得了睡莲类植物的2个ITS系统树, 结果也支持现有分子系统学研究中有关金鱼藻处于较原始位置的观点.Abstract: Water-lily plants are key group on study of the flowering plant phylogeny. The internal transcribed spacers (ITS) and the 5.8S coding region of nuclear ribosomal DNA (nrDNA) of seven taxa were sequenced to study molecular phylogeny and evolution of Nymphaeales sensu lato. The ITS phylogenetic trees were constructed based on the sequences and a combined data set with sequences from GenBank. The results obtained from ITS sequences also supported that the genus Ceratophyllum could be considered the ancestral group in the order, as the results of most molecular systematic studies.
-
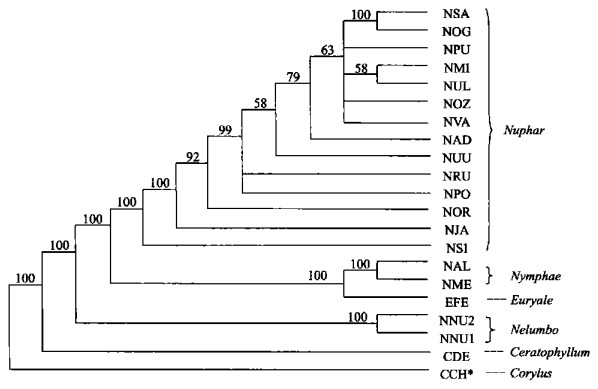
图 1 用简约法和邻接法构建的ITS系统树
数字表示各分支自展数据支持率; *表示外类群, 分类群缩写见表 1, 左为MP, 右为NJ
Fig. 1. ITS phylogenetic trees using maximum parsimony and neighbor-joining methods
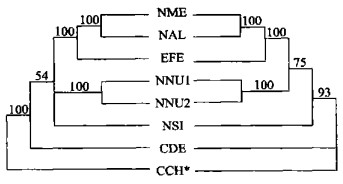
图 2 用简约法构建的ITS系统树(组合数据集)
数字表示各分支自展数据支持率; *表示外类群; 分类群缩写见表 1; 右边斜体为属名
Fig. 2. ITS phylogenetic trees using maximum parsimony (combined data set)
表 1 实验材料的种名、缩写及来源
Table 1. Species, abbreviation and source

-
[1] 杨群. 古生物学领域的新辟园地——分子古生物研究[J]. 古生物学报, 1995, 34(3): 265-276. https://www.cnki.com.cn/Article/CJFDTOTAL-GSWX199503000.htmYANG Q. A frontal area in palaeontology—molecular fossil studies[J]. Acta Palaeontologica Sinica, 1995, 34(3): 265 -276. https://www.cnki.com.cn/Article/CJFDTOTAL-GSWX199503000.htm [2] 杨洪, 程安进, 杨群. 地质体中主要生物分子的研究方法及应用[J]. 地质论评, 1998, 44(1): 44-51. doi: 10.3321/j.issn:0371-5736.1998.01.007YANG H, CHENG A J, YANG Q. Research method and application of major biology molecular in geology[J]. Geology Review, 1998, 44(1): 44-51. doi: 10.3321/j.issn:0371-5736.1998.01.007 [3] Qiu Y L, Lee J, Bernasconi Q F, et al. The earliest angiosperms: evidence from mitochondrial, plastid and nuclear genomes[J]. Nature, 1999, 402: 404-407. doi: 10.1038/46536 [4] Mathews S, Michael J D. The root of angiosperm phylogeny inferred from duplicate phytochrome genes[J]. Science, 1999, 286: 947-950. doi: 10.1126/science.286.5441.947 [5] Barkman T J, Chenery G, Mcneal J R, et al. Independent and combined analyses of sequences from all three genomic compartments converge on the root of flowering plant phylogeny[J]. Proc Natl Acad Sci, 2000, 97: 13166-13171. doi: 10.1073/pnas.220427497 [6] 侯宽昭, 吴德邻, 高蕴璋, 等. 中国种子植物科属词典(修订版)[M]. 北京: 科学出版社, 1984.HOU K Z, WU D L, GAO Y Z, et al. Dictionary of spermatophyte classify in China[M]. Beijing: Science Press, 1984. [7] 中国科学院武汉植物研究所. 中国水生微管束植物图谱[M]. 武汉: 湖北人民出版社, 1980.Wuhan Institute of Botany, Chinese Academy of Sciences. A collection of hydrophily canaliculus plant in China[M]. Wuhan: Hubei People Press, 1980. [8] 钟扬, 张晓艳, 黄德世. 睡莲目的数量分支分类学研究[J]. 生物数学学报, 1990, 5(2): 156-161. https://www.cnki.com.cn/Article/CJFDTOTAL-SWSX199002009.htmZHONG Y, ZHANG X Y, HUANG D S. Study of quantity cladistics on water-lilies[J]. Acta Biomathematics Sinica, 1990, 5(2): 156-161. https://www.cnki.com.cn/Article/CJFDTOTAL-SWSX199002009.htm [9] White T J, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics [A]. In: Innis M, Gelfand D, Sninskr J, et al, eds. PCR protocols: a guide to methods and applications[C]. San Diego: Academic Press, 1990. 315-322. [10] Baldwin B G. Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the Compositae[J]. Molec Phylogen Evol, 1992, 1: 3-16. doi: 10.1016/1055-7903(92)90030-K [11] Baldwin B G, Sanderson M J, Porter J M, et al. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny[J]. Ann Missouri Bot Gard, 1995, 82: 247-277. doi: 10.2307/2399880 [12] Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment[J]. Comput Appl Biosci, 1992, 8: 189-191. [13] Felsenstein J. PHYLIP(phylogeny inference package)version 3.572 c[CP/DK]. Department of Genetics, University of Washington, Seattle, 1996. [14] 唐先华. 睡莲类植物的nrDNA ITS序列分析及分子系统发育与进化研究[D]. 武汉: 中国科学院植物研究所, 1998.TANG X H. Molecular phylogeny and evolution of water-lilies based onn rDNA ITS sequences[D]. Wuhan: Wuhan Institute of Botany, Chinese Academy of Sciences, 1998. [15] Doyle J J, Doyle J L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue[J]. Phytochem Bull, 1987, 19: 11-15. [16] Wen J, Zimmer E A. Phylogeny and biogeograph of Panax L(the ginseng genus, Araliaceae): inferences from ITS sequences of nuclear ribosomal DNA[J]. Mol Phylogenet Evol, 1996, 6: 166-177. [17] Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap[J]. Evolution, 1985, 39: 783 -791. doi: 10.1111/j.1558-5646.1985.tb00420.x [18] Felsentein J. Evolutionary trees from DNA sequences: a maximum-likelihood approach[J]. J Mol Evol, 1981, 17: 368-376. doi: 10.1007/BF01734359 [19] Jukes T H, Cantor C R. Evolution of protein molecules [A]. In: Munro HN, ed. Mammalian protein metabolism [C]. New York: Academic Press, 1969. 21-132. [20] Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees[J]. Mol Miol Evol, 1987, 4: 406-425. doi: 10.1093/oxfordjournals.molbev.a040454 [21] Les D H, Garvin D K, Wimpee C F. Molecular evolutionary history of ancient aquatic angiosperms[J]. Proc Natl Acad Sci, 1991, 88: 10119-10123. doi: 10.1073/pnas.88.22.10119 [22] Chase M W, Balthazar M V, Hoot S B, et al. Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbc l[J]. Ann Missouri Bot Gard, 1993, 80: 528-580. doi: 10.2307/2399846 [23] Ito M. Phylogenetic systematics of the Nymphaeales[J]. Bot Mag Tokyo, 1987, 100: 17-35. doi: 10.1007/BF02488417 -








 下载:
下载: